Tokyo, 24 September, /AJMEDIA/
As people around the world marveled in July at the most detailed pictures of the cosmos snapped by the James Webb Space Telescope, biologists got their first glimpses of a different set of images — ones that could help revolutionize life sciences research.
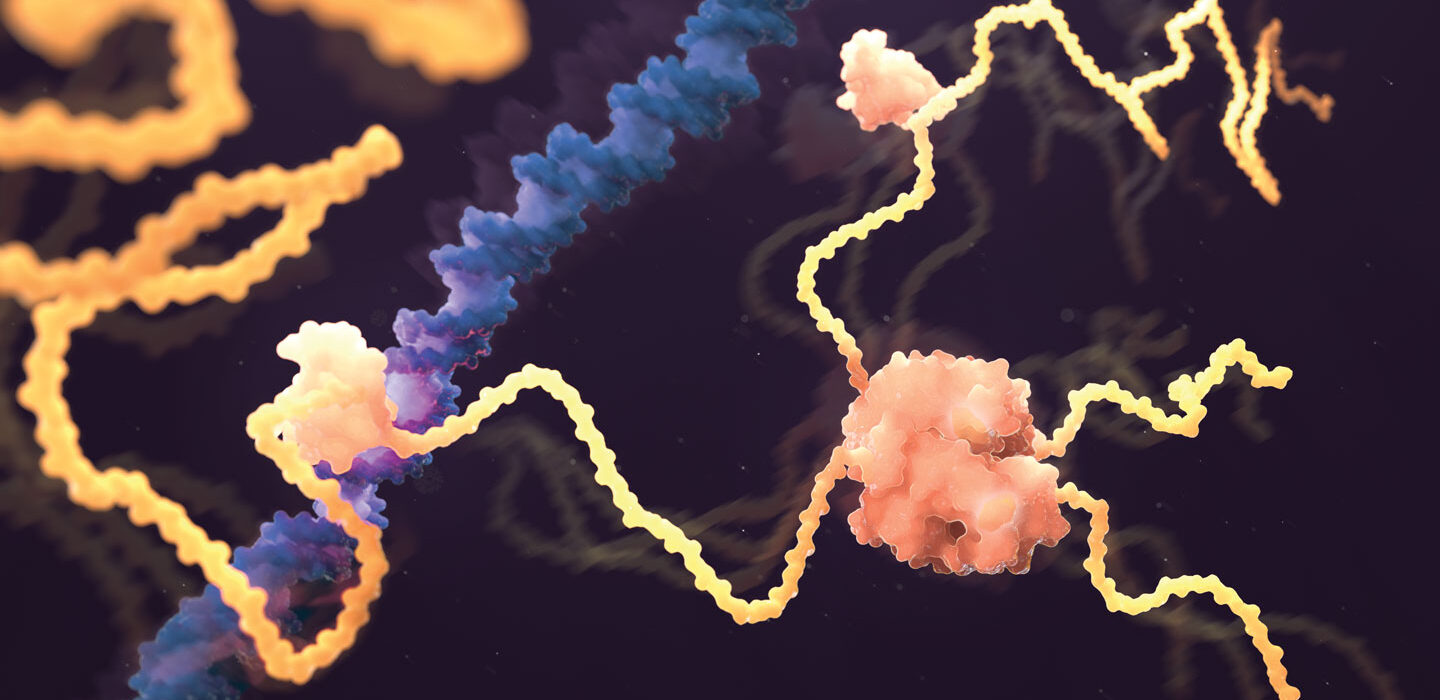
The images are the predicted 3-D shapes of more than 200 million proteins, rendered by an artificial intelligence system called AlphaFold. “You can think of it as covering the entire protein universe,” said Demis Hassabis at a July 26 news briefing. Hassabis is cofounder and CEO of DeepMind, the London-based company that created the system. Combining several deep-learning techniques, the computer program is trained to predict protein shapes by recognizing patterns in structures that have already been solved through decades of experimental work using electron microscopes and other methods.
The AI’s first splash came in 2021, with predictions for 350,000 protein structures — including almost all known human proteins. DeepMind partnered with the European Bioinformatics Institute of the European Molecular Biology Laboratory to make the structures available in a public database.
July’s massive new release expanded the library to “almost every organism on the planet that has had its genome sequenced,” Hassabis said. “You can look up a 3-D structure of a protein almost as easily as doing a key word Google search.”
These are predictions, not actual structures. Yet researchers have used some of the 2021 predictions to develop potential new malaria vaccines, improve understanding of Parkinson’s disease, work out how to protect honeybee health, gain insight into human evolution and more. DeepMind has also focused AlphaFold on neglected tropical diseases, including Chagas disease and leishmaniasis, which can be debilitating or lethal if left untreated.
The release of the vast dataset was greeted with excitement by many scientists. But others worry that researchers will take the predicted structures as the true shapes of proteins. There are still things AlphaFold can’t do — and wasn’t designed to do — that need to be tackled before the protein cosmos completely comes into focus.
Having the new catalog open to everyone is “a huge benefit,” says Julie Forman-Kay, a protein biophysicist at the Hospital for Sick Children and the University of Toronto. In many cases, AlphaFold and RoseTTAFold, another AI researchers are excited about, predict shapes that match up well with protein profiles from experiments. But, she cautions, “it’s not that way across the board.”
Predictions are more accurate for some proteins than for others. Erroneous predictions could leave some scientists thinking they understand how a protein works when really, they don’t. Painstaking experiments remain crucial to understanding how proteins fold, Forman-Kay says. “There’s this sense now that people don’t have to do experimental structure determination, which is not true.”